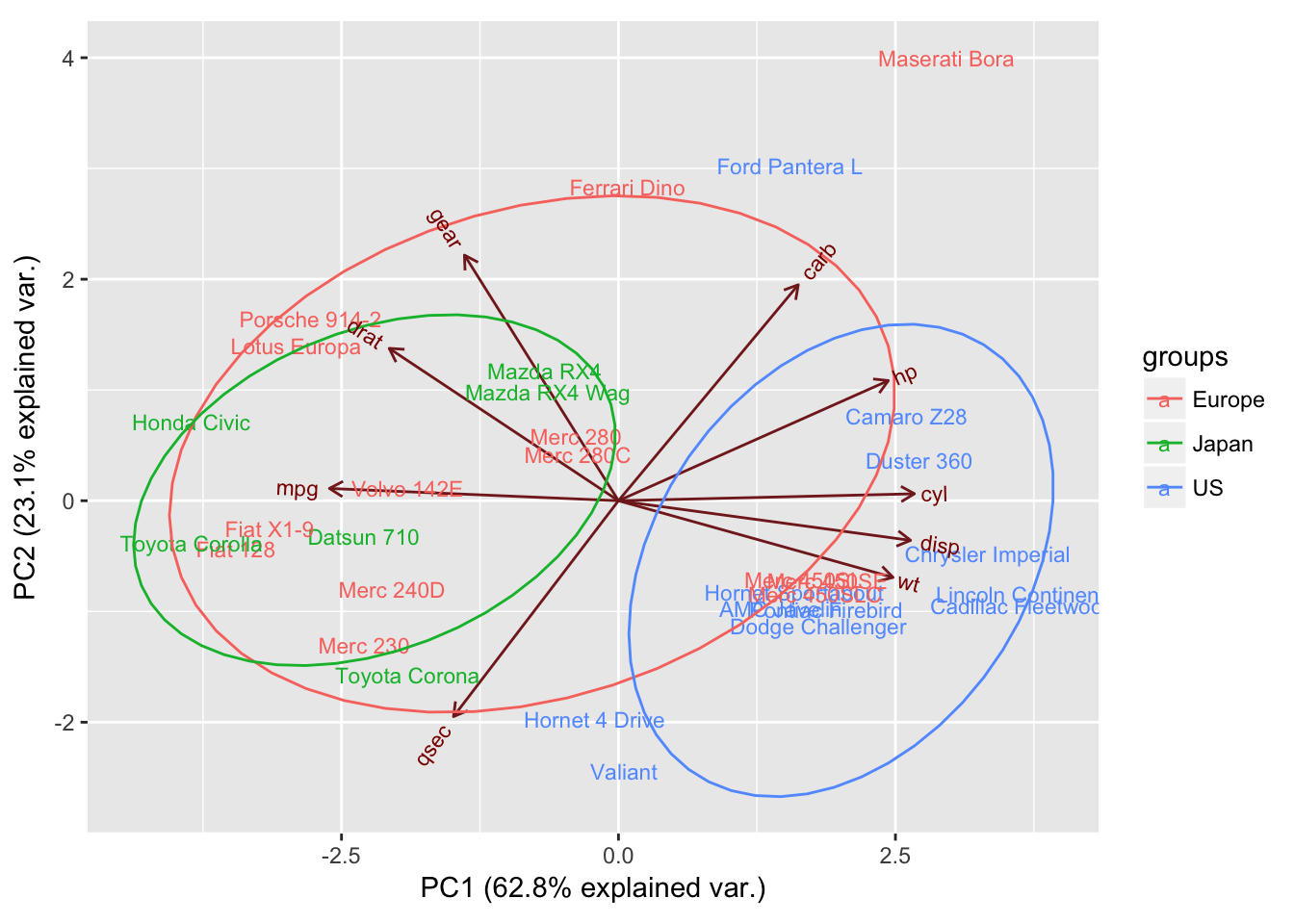
Principal component analysis (PCA) biplot generated in R using... Download Scientific Diagram
Principal Component Analysis (PCA) is a widely-used statistical technique in the field of data science and machine learning. This article provides a step-by-step guide on implementing PCA in R, a popular programming language among statisticians and data analysts.
GraphPad Prism 10 Statistics Guide Graphs for Principal Component Analysis
PCA is a multivariate technique that is used to reduce the dimension of a data set. More precisely, PCA is concerned with explaining the variance -covariance structure through a few linear combinations of the original variables.
Principal Component Analysis in R vs Articles STHDA
Introduction: Principal component analysis (PCA) is a common technique for performing dimensionality reduction on multivariate data. By transforming the data into principal components, PCA allows.

Principal Component Analysis (PCA) 101, using R Towards Data Science
PCA is used in exploratory data analysis and for making decisions in predictive models. PCA commonly used for dimensionality reduction by using each data point onto only the first few principal components (most cases first and second dimensions) to obtain lower-dimensional data while keeping as much of the data's variation as possible.
PCA Principal Component Analysis Essentials Articles STHDA
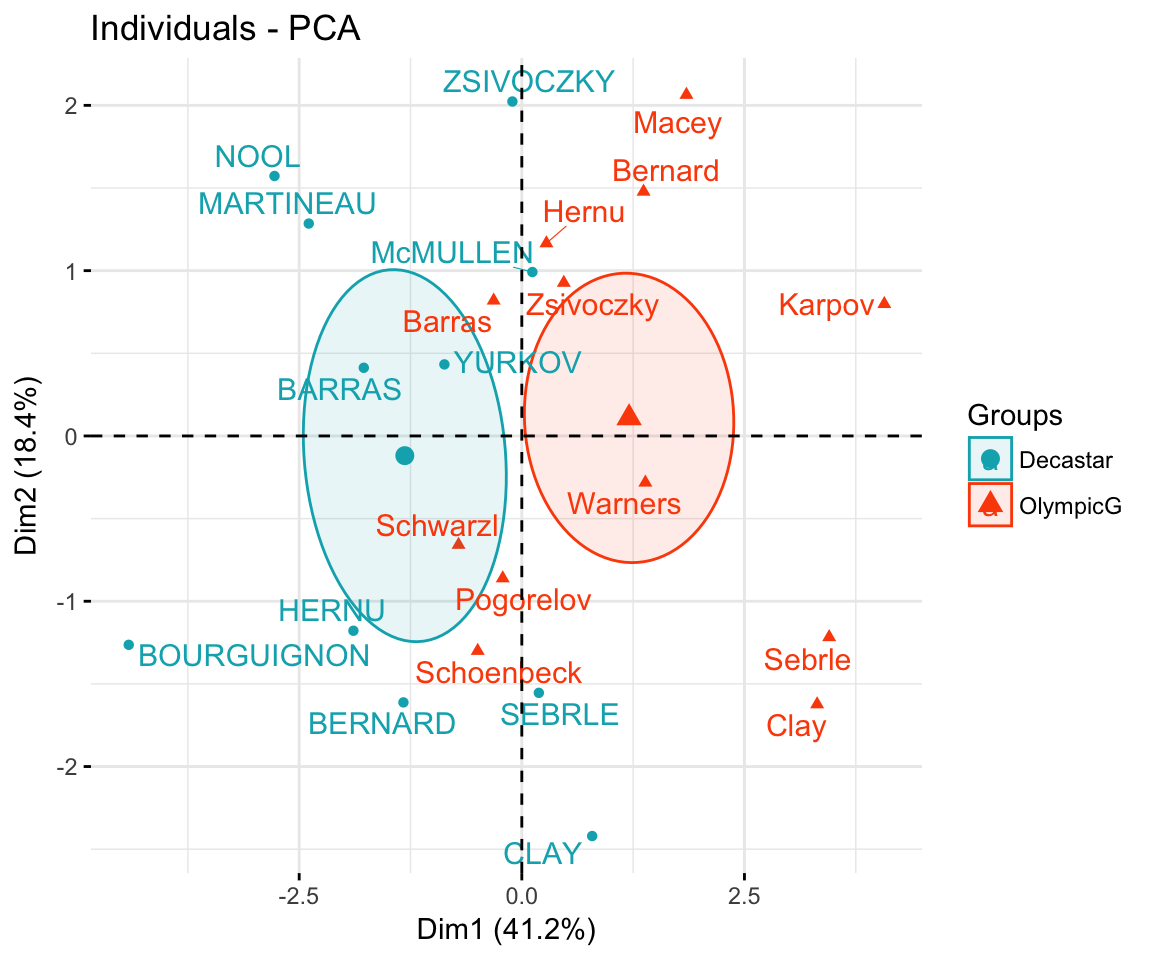
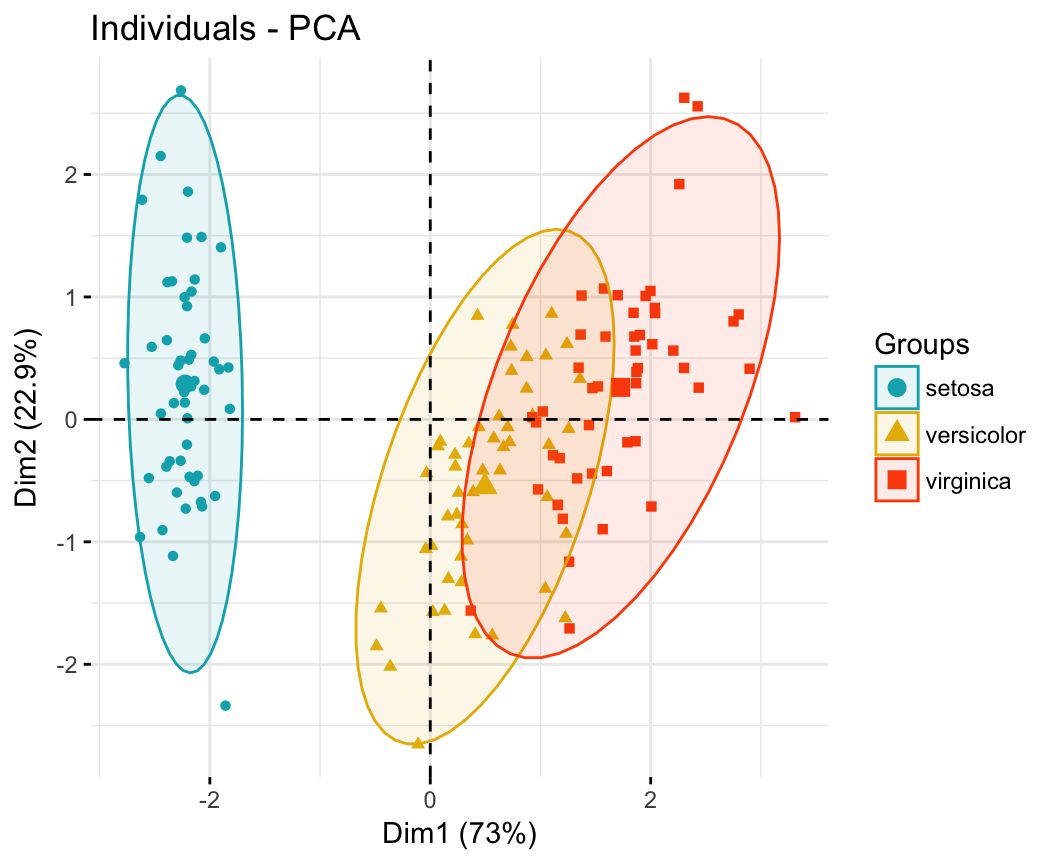
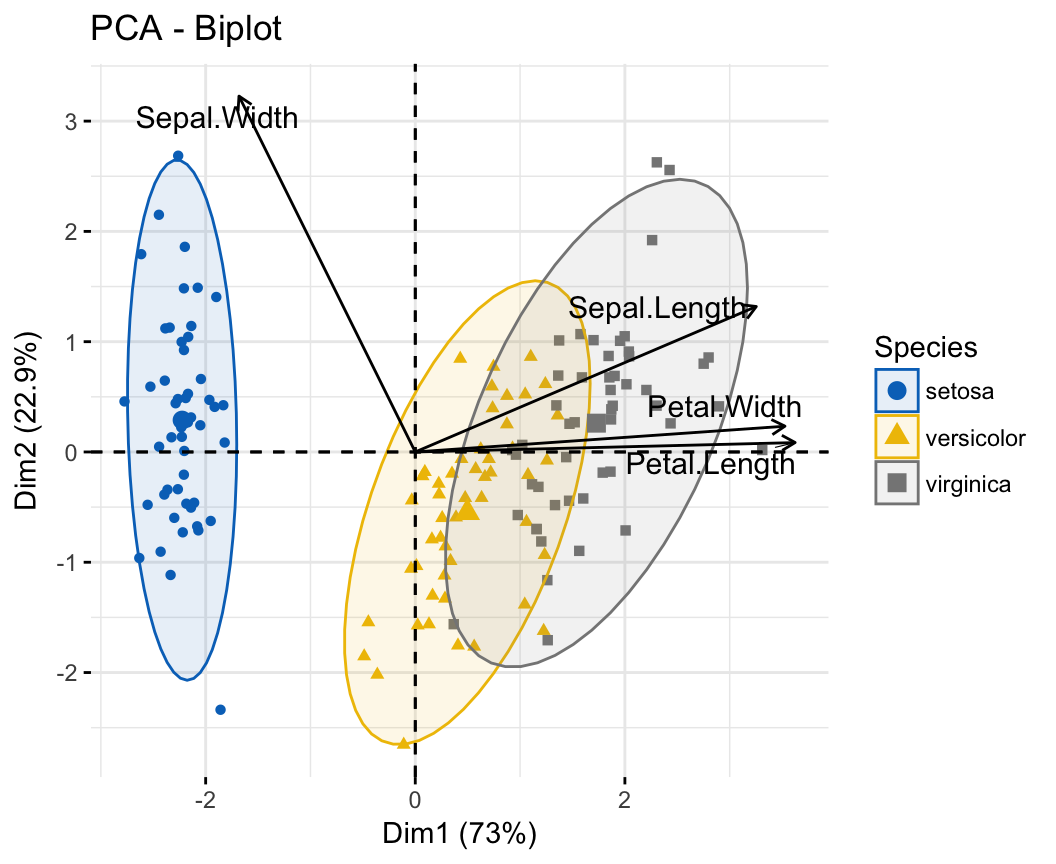
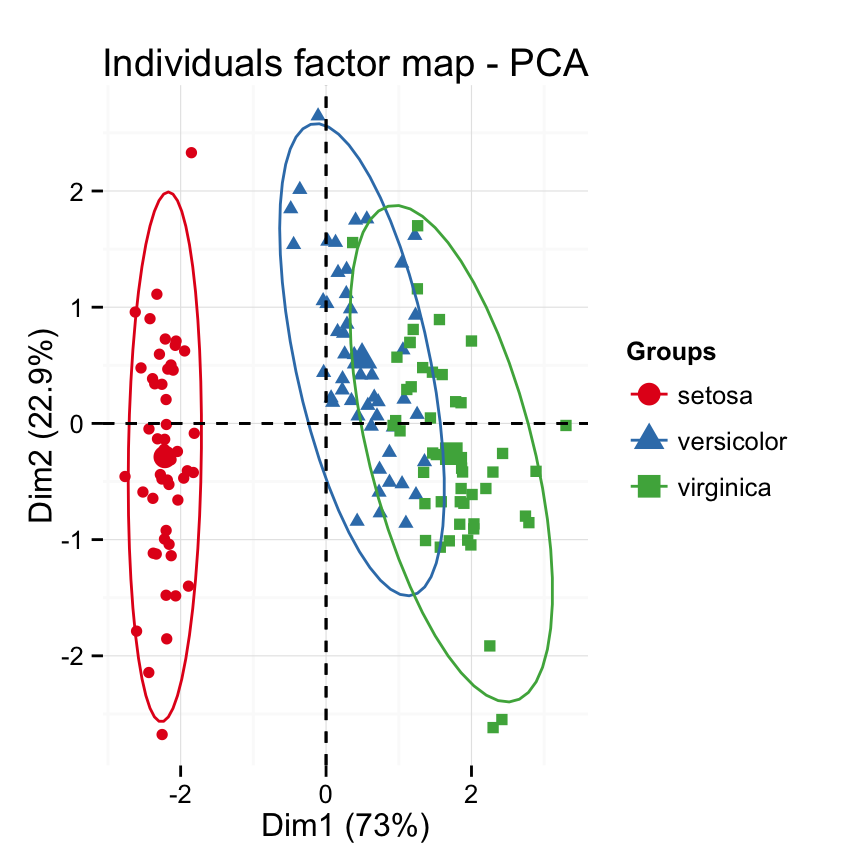
Case 1: Continuous variables. In the situation where you have a multidimensional data set containing multiple continuous variables, the principal component analysis (PCA) can be used to reduce the dimension of the data into few continuous variables containing the most important information in the data. Next, you can perform cluster analysis on the PCA results.
PCA Principal Component Analysis Essentials Articles STHDA
In this tutorial you'll learn how to perform a Principal Component Analysis (PCA) in R. The table of content is structured as follows: 1) Example Data & Add-On Packages 2) Step 1: Calculate Principal Components 3) Step 2: Ideal Number of Components 4) Step 3: Interpret Results 5) Video, Further Resources & Summary
Principal component analysis (PCA) in R Rbloggers
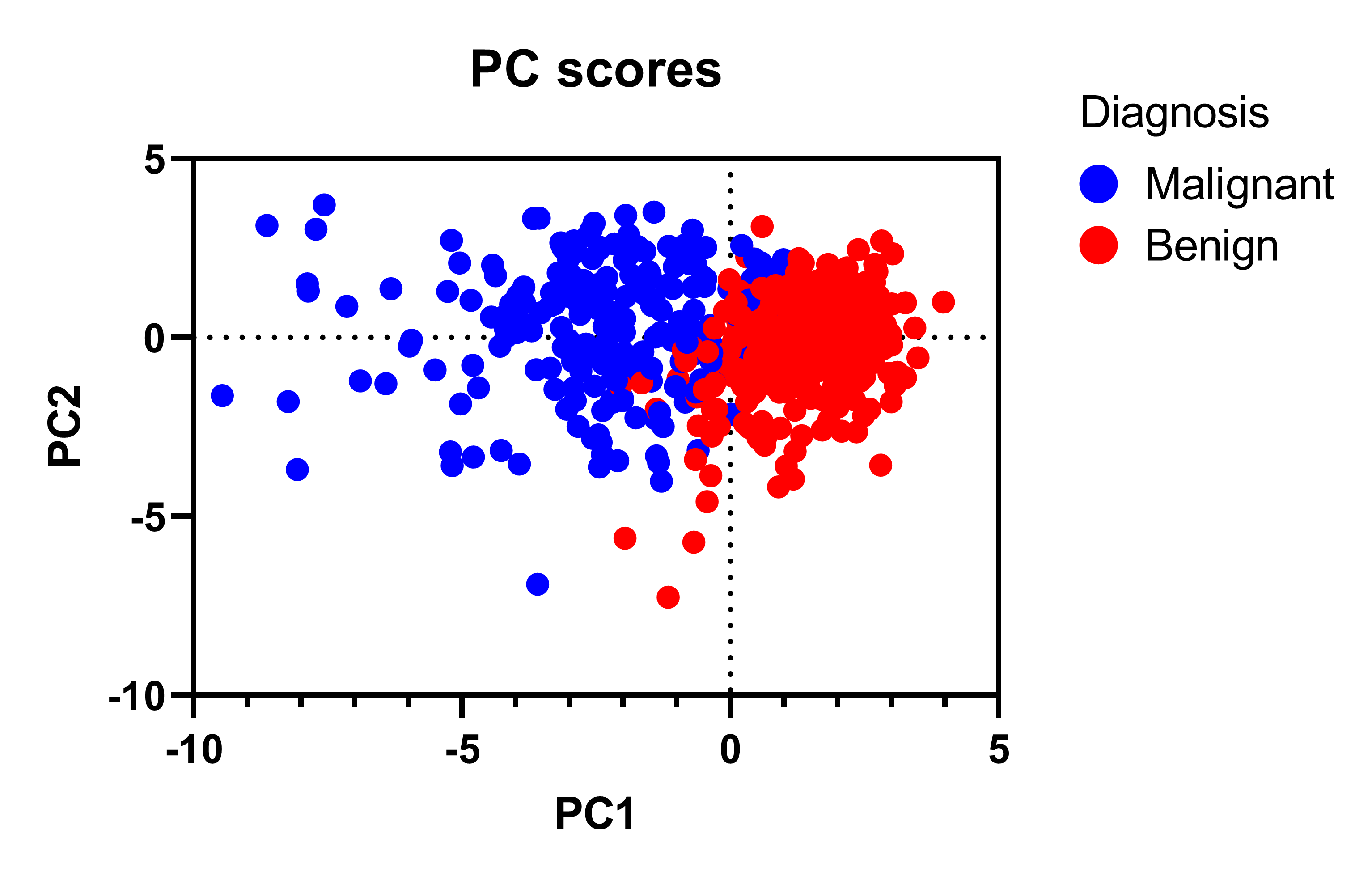
Principal Component Analysis (PCA) 101, using R Peter Nistrup · Follow Published in Towards Data Science · 8 min read · Jan 29, 2019 2 Improving predictability and classification one dimension at a time! "Visualize" 30 dimensions using a 2D-plot! Basic 2D PCA-plot showing clustering of "Benign" and "Malignant" tumors across 30 features.

Principal Component Analysis (PCA) in R YouTube
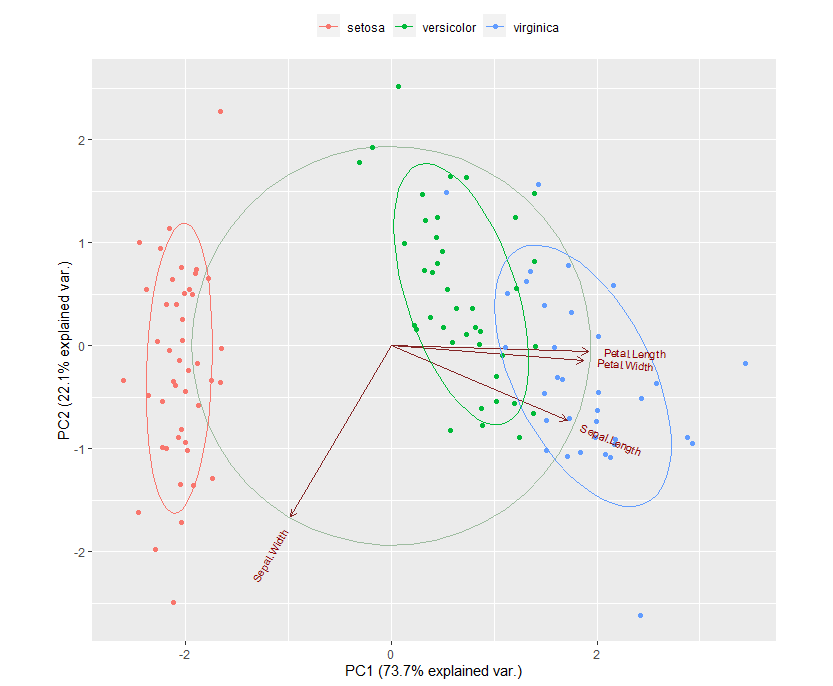
Principal component analysis ( PCA) allows us to summarize and to visualize the information in a data set containing individuals/observations described by multiple inter-correlated quantitative variables. Each variable could be considered as a different dimension.
Principal component analysis in R vs. R software and data mining Easy
This R tutorial describes how to perform a Principal Component Analysis ( PCA) using the built-in R functions prcomp () and princomp (). You will learn how to predict new individuals and variables coordinates using PCA. We'll also provide the theory behind PCA results.
R PCA Tutorial (Principal Component Analysis) DataCamp
For many or most types of analysis, one would just do the first three steps, which provides the scores and loadings that are usually the main result of interest. In some cases,. 2There are other functions in R for carrying out PCA. See the PCA Functions vignette for the details. 5. Fe2O3 Cu centered & scaled values −1 0 1 2
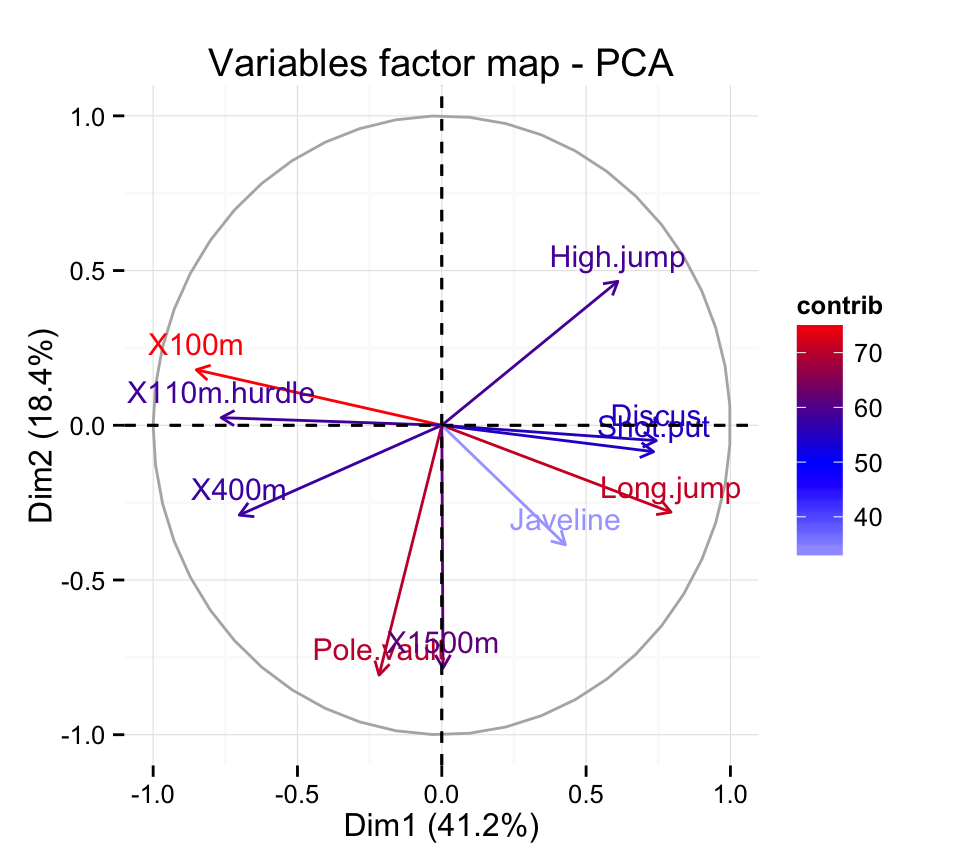
fviz_pca Quick Principal Component Analysis data visualization R software and data mining
A principal component analysis of the data can be applied using the prcomp function. The result is a list containing the coefficients defining each compo-nent (sometimes referred to as loadings), the principal component scores, etc. The required code is (omitting the scorevariable)
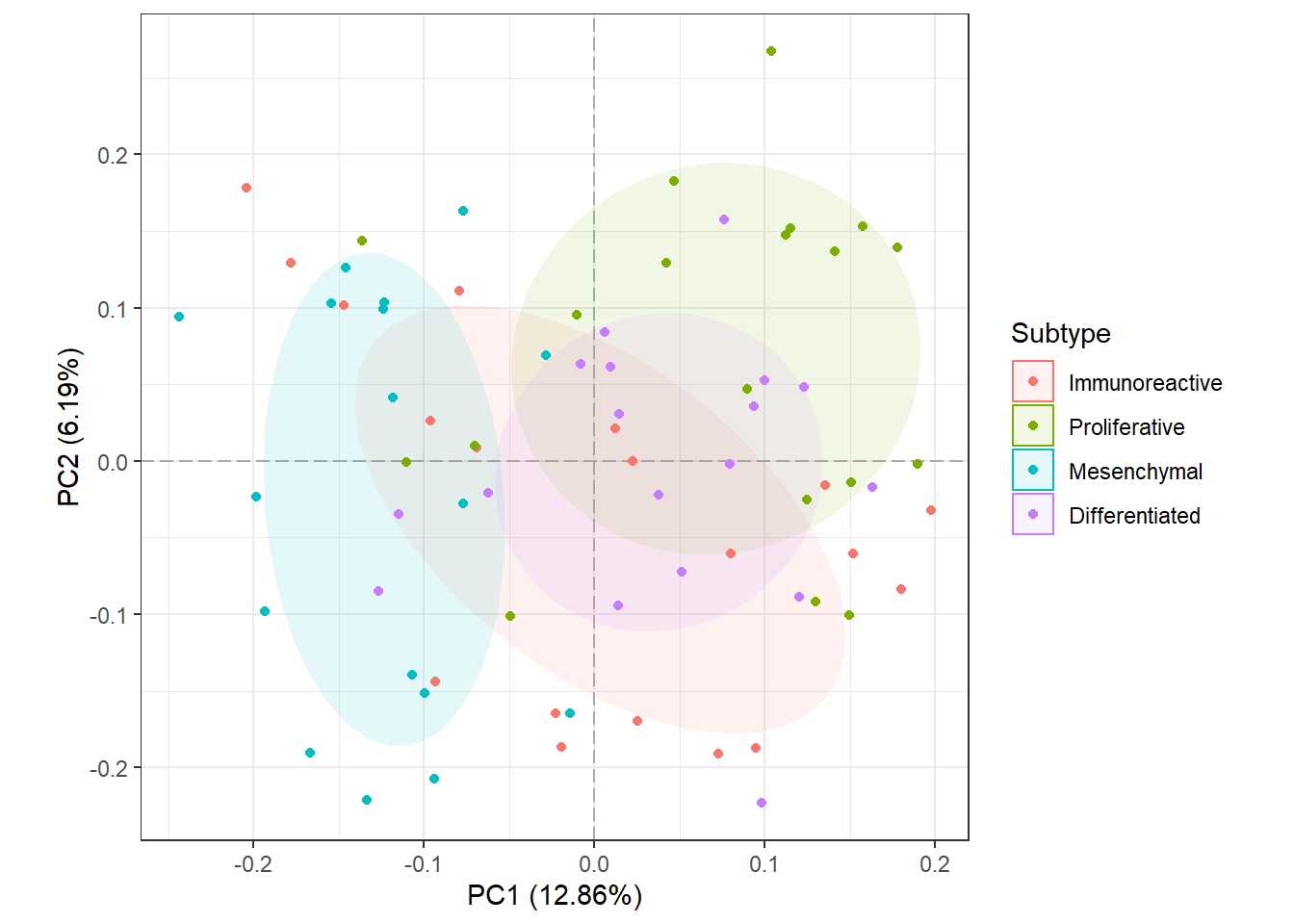
5.4 PCA Proteomics Data Analysis in R/Bioconductor
Principal components analysis, often abbreviated PCA, is an unsupervised machine learning technique that seeks to find principal components - linear combinations of the original predictors - that explain a large portion of the variation in a dataset.
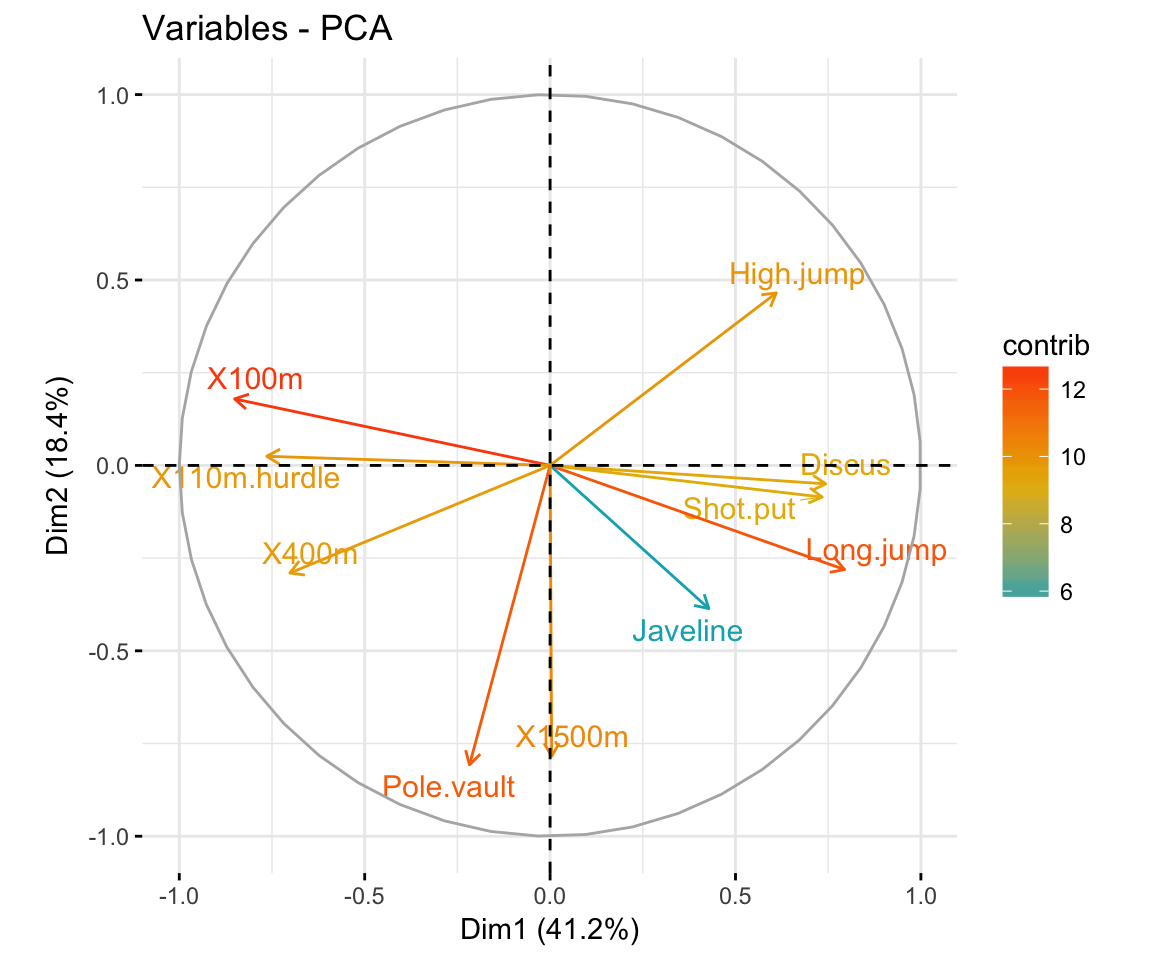
Principal Component Analysis in R vs Articles STHDA
Feb 15, 2018. Principal Component Analysis (PCA) is unsupervised learning technique and it is used to reduce the dimension of the data with minimum loss of information. PCA is used in an application like face recognition and image compression. PCA transforms the feature from original space to a new feature space to increase the separation.
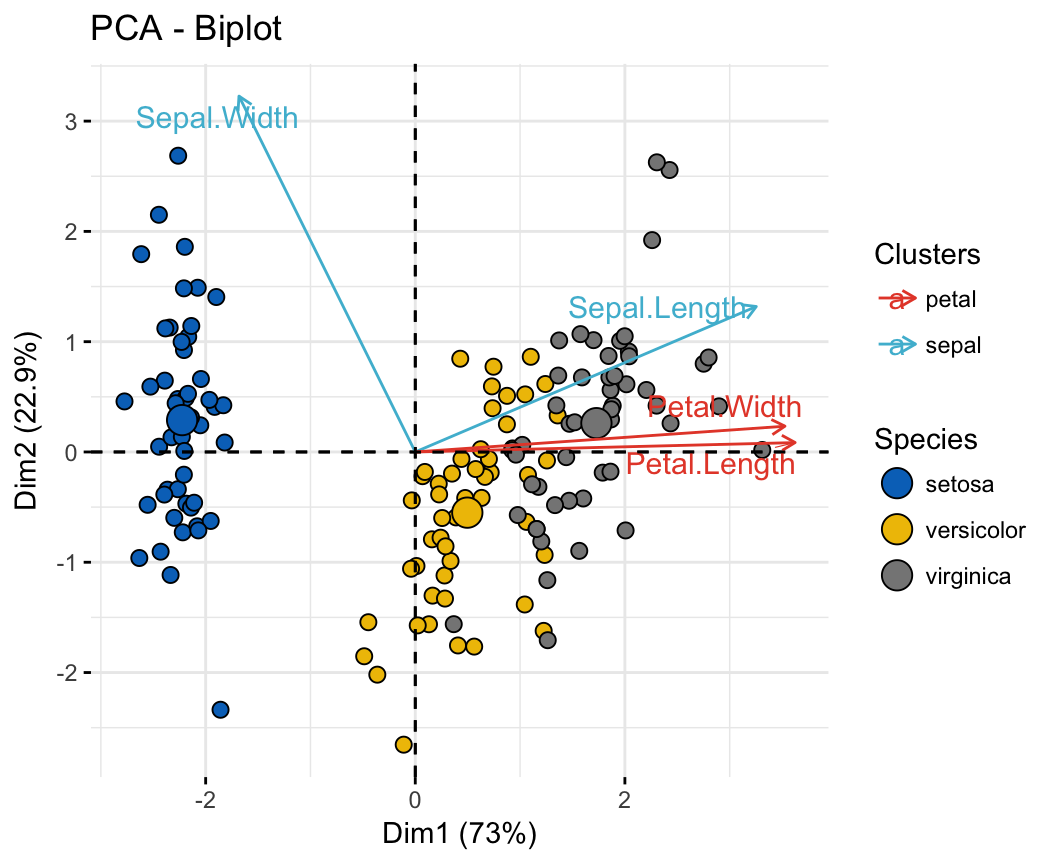
PCA Principal Component Analysis Essentials Articles (2023)
Plotting PCA (Principal Component Analysis) {ggfortify} let {ggplot2} know how to interpret PCA objects. After loading {ggfortify}, you can use ggplot2::autoplot function for stats::prcomp and stats::princomp objects. library(ggfortify) df <- iris[1:4] pca_res <- prcomp(df, scale. = TRUE) autoplot(pca_res)
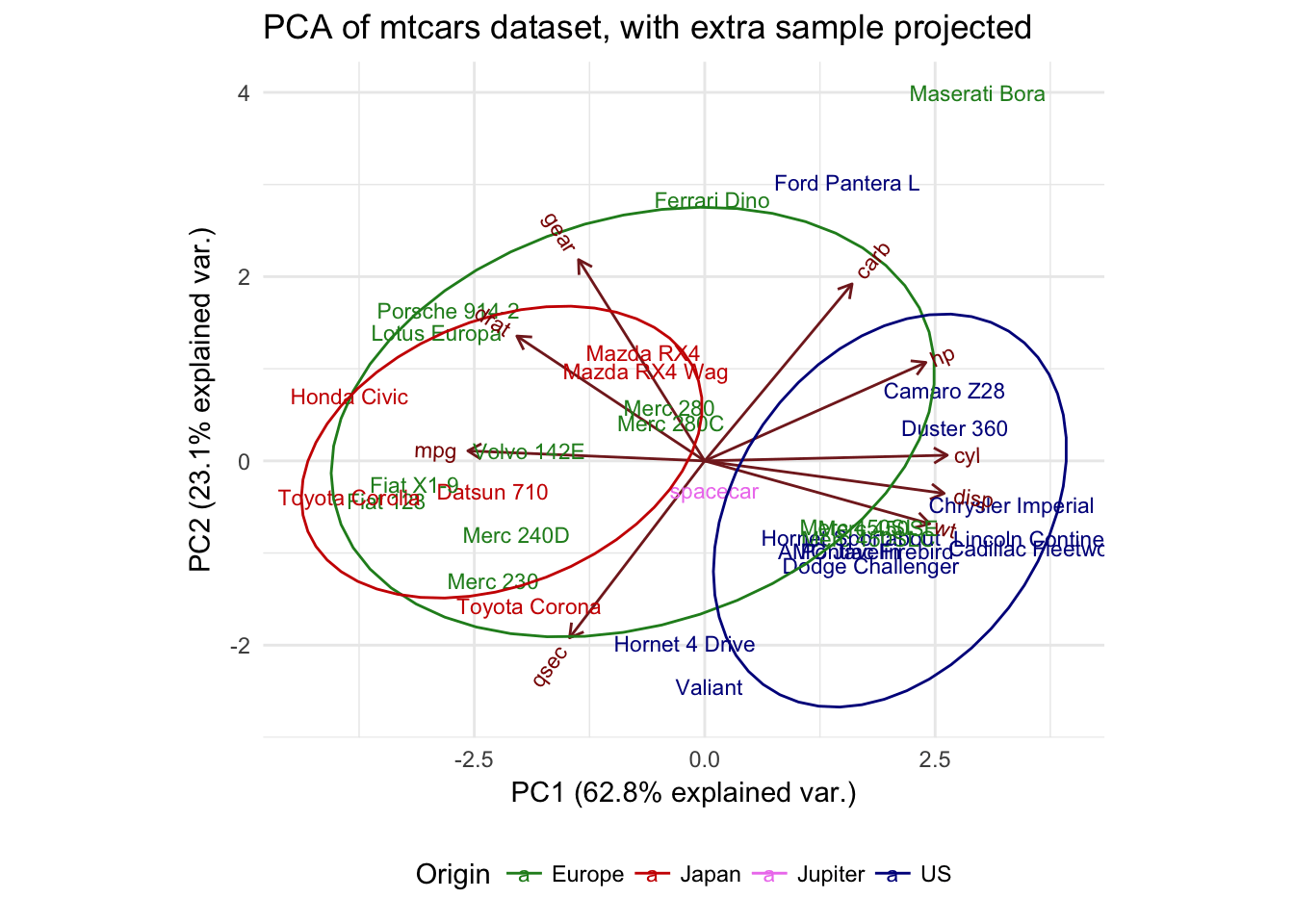
R PCA Tutorial (Principal Component Analysis) DataCamp
PCA is commonly used as one step in a series of analyses. You can use PCA to reduce the number of variables and avoid multicollinearity, or when you have too many predictors relative to the number of observations. tl;dr This tutorial serves as an introduction to Principal Component Analysis (PCA). 1
Apply Principal Component Analysis in R (PCA Example & Results)
PCA is an exploratory data analysis based in dimensions reduction. The general idea is to reduce the dataset to have fewer dimensions and at the same time preserve as much information as possible.